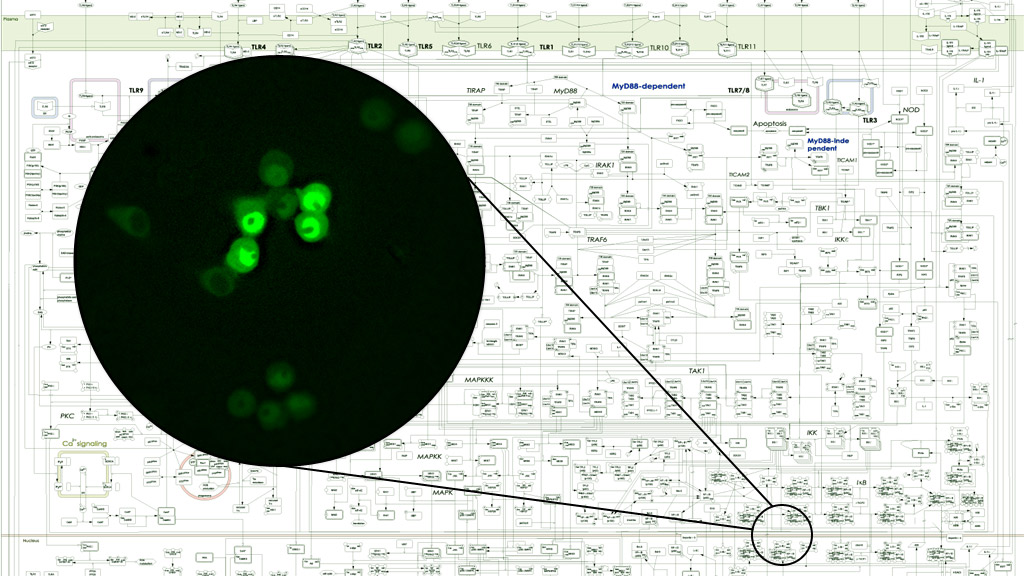
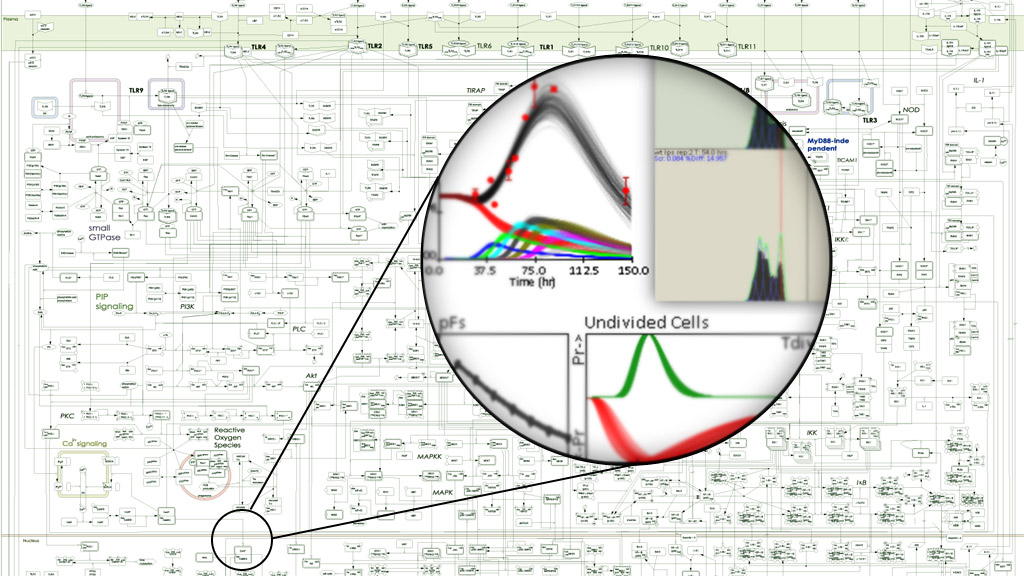
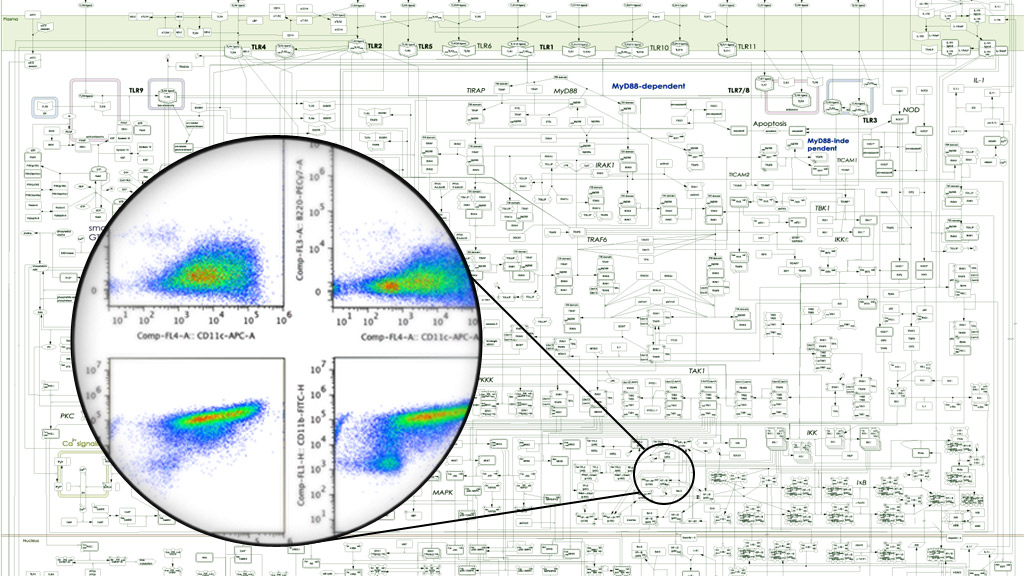
We aim to develop a predictive understanding of the Signaling Systems that control immune responses. Projects focus on the stimulus-specific responses of immune sentinel macrophages, antibody responses produced by B-cells, and the impact of immune responses on the generation of new immune cells via hematopoiesis. The Laboratory integrates a variety of traditional and cutting-edge experimental approaches with various computational methods. Our researchers come from diverse backgrounds and education, but we all study the intricate and powerful systems activated when immune cells are exposed to pathogens.
Immune responses are incredibly dynamic and multi-scale, involving regulation at the biophysical scale of single ligand-protein binding to the high level interactions between numerous cells in immune organs. Multicellular organisms coordinate these responses to reliably deal with the constant threat of invasive bacteria, viruses, protists, and parasites.
Using experimental approaches of biochemistry, molecular biology, fluorescence microscopy and next generation sequencing, in conjunction with cutting-edge bioinformatics, data-driven computational modeling, and knowledge-based mathematical modeling, we aim to connect top-down “unbiased” Systems Biology with and bottom-up “hypothesis-driven” Systems Biology.
Our laboratory draws talents from diverse cultural, ethnic, and racial backgrounds. As researchers and educators, we recognize that we make an impact not only through our scientific pursuit but also our influence on society. We embrace the opportunities of living in the diverse metropolis of Los Angeles by engaging in science education in local schools.