

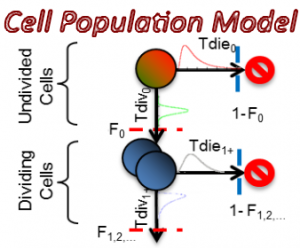
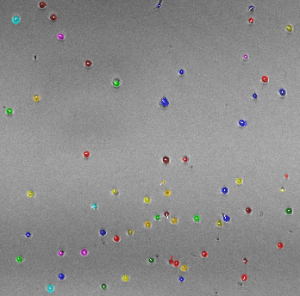
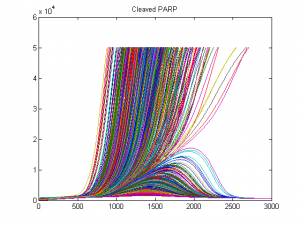
I am interested in understanding how cell population dynamics are coordinated on the cellular and biochemical levels. A prime example, the mammalian immune response involves seemingly stochastic cellular decisions to respond, divide, and undergo programmed cell death. Understand how immune cells make decisions that result in an effective population response sheds light on the nature of multi-cellular biology and may provide insights into treating immune disorders and cancers. To this end, I have developed computational tools for describing cell population dynamics in terms of stochastic cellular processes, cell tracking software for quantifying time-lapse microscopy movies, performed single-cell RNAseq experiments, ran immunfluorescence studies, and constructed kinetic ODE models which incorporate the cell-cycle, apoptosis, and NFkB signaling. This has allowed me to characterize the responses of primary mouse B cells under diverse conditions in a multi-scale manner.
 dnrios@nullucsd.edu
dnrios@nullucsd.edu
Office: 858-822-4673
B.S. Biochemistry, California Polytechnic State University San Luis Obispo, 2008
I am interested in the dynamics of interferon beta production in response to TLR stimulation. I plan to examine, through experimental and computational techniques, the coordination of NF-kappaB and ISGF3 in mounting an innate immune response.
 -)
-)ploriaux@nullbioinf.ucsd.edu
Office: NSB 4318
Phone: 858-822-4673
B.S. Bioengineering, Univ. of Washington 2001
M.S. Computer Science & Engineering, Univ. of California San Diego, 2011
My current research interests are in understanding the molecular mechanisms that process information in living cells. Cell systems represent a particular challenge in this regard in that their machinery is capable of operating over many orders of magnitude in both space and time. Computational modeling is particularly suited to making conjectures about how systems operate over a high dynamic range in these domains, and these conjectures can then be verified or refuted in the laboratory. Presently I am constructing a model of concurrent activation of the JNK and NF-kappa B signaling pathways, with the goal of understanding how the basal state of the system biases the cellular response towards one or the other of these two contradictory signals. The results could have implications to our basic understanding of why different cell types respond differently to the same set of inflammatory and apoptotic signals.
| Citation | Link |
|---|---|
| Of elections and cell-death decisions. Loriaux P, Hoffmann A. Mol Cell. 2009 May 15;34(3):257-8. |
PubMed |
| A framework for modeling the relationship between cellular steady-state and stimulus-responsiveness. Loriaux PM, Hoffmann A. Methods Cell Biol. 2012;110:81-109. |
PubMed |
| Characterizing the Relationship between Steady State and Response Using Analytical Expressions for the Steady States of Mass Action Models. Loriaux PM, Tesler G, Hoffmann A. PLoS Comput Biol. 2013 Feb;9(2):e1002901. |
PubMed |
| A protein turnover signaling motif controls the stimulus-sensitivity of stress response pathways. Loriaux PM, Hoffmann A. PLoS Comput Biol. 2013 Feb;9(2):e1002932. |
PubMed |
 j0huang@nullucsd.edu
j0huang@nullucsd.edu
Office: NSB 3320
Phone: 858-822-4673
B.S. Chemistry, Cornell, 2007
My research interests are to study the pleiotropic effects of drugs on the dynamic regulation of NFkB signaling system using both biochemical assays and single cell experiments. Using a systems pharmacology approach, the goal is to determine a combination of chemical perturbations which could yield specific target effects on gene expression.
| Citation | Link |
|---|---|
| Control of self-assembly of lithographically patternable block copolymer films. Bosworth JK, Paik MY, Ruiz R, Schwartz EL, Huang JQ, et al ACS Nano. 2008 Jul;2(7):1396-402. |
PubMed |

B.S. Biology and B.S. Molecular and Microbiology
University of Central Florida 2007
I am examining the effects of different bacterial pathogens on the dynamics and kinetics of the NF-kappaB signaling network. Through these investigations I hope to learn more about how pathogens might alter or disrupt host cell signaling and how different pathogens might elicit different immune responses.
Cheng CS, Feldman KE, et al. The Specificity of Innate Immune Responses Is Enforced by Repression of Interferon Response Elements by NF-B p50. 22 February 2011. Science Signaling. 4 (161), ra11.

B.A. in Molecular Biology, Princeton University 2004
Fellow, National Science Foundation Graduate Research Program
I am interested in the coupling between transcriptional elongation and splicing. In addition to modeling, I am pursuing a high-throughput assay of splicing events in pathogen response genes.
| Citation | Link |
|---|---|
| Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Shih VF, Davis-Turak J, Macal M, Huang JQ, Ponomarenko J, Kearns JD, Yu T, Fagerlund R, Asagiri M, Zuniga EI, Hoffmann A. Nat Immunol. 2012 Dec;13(12):1162-70. |
PubMed |
| Human-specific transcriptional networks in the brain. Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, Geschwind DH. Neuron. 2012 Aug 23;75(4):601-17 |
PubMed |
| Transcriptional architecture of the primate neocortex. Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MM, Serikawa K, Lemon T, Morgan R, Copeland C, Smith K, Cullen V, Davis-Turak J, Lee CK, Sunkin SM, Loboda AP, Levine DM, Stone DJ, Hawrylycz MJ, Roberts CJ, Jones AR, Geschwind DH, Lein ES. Neuron. 2012 Mar 22;73(6):1083-99 |
PubMed |
| A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. Genomics. 2011 Oct;98(4):266-71 |
PubMed |
| Tauopathy with paired helical filaments in an aged chimpanzee. Rosen RF, Farberg AS, Gearing M, Dooyema J, Long PM, Anderson DC, Davis-Turak J, Coppola G, Geschwind DH, Par JF, Duong TQ, Hopkins WD, Preuss TM, Walker LC. J Comp Neurol. 2008 Jul 20;509(3):259-70. |
PubMed |
 Chemistry and Biochemistry Graduate Student (2008-)
Chemistry and Biochemistry Graduate Student (2008-)
B.S. Chemistry, Biochemistry,Seattle Pacific University, 2008
I am interested in the mechanisms of IKK activation and regulation in NFκB signaling.

B.S. Biochemistry and Cell Biology, UCSD
B-cells undergo dramatic expansion during immune response, which involves one of the most rapid proliferation rates known for mammalian cells. B-cell survival and death is highly regulated, and the proliferative program is limited to about 12 divisions.
The NF-kB signaling system plays a critical role in B-cell biology. Two distinct NF-kB activation pathways have been described to induce overlapping sets of NF-kB transcription factors containing the three activation domain containing NF-kB proteins, RelA, RelB, and cRel. My research is focused on understanding which NF-kB dimers control B-cell survival and proliferation. This study is complicated by the overlap of these two B-cell processes, as well as the interdependencies of NF-kB dimers produced by the NF-kB signaling system in response to B-cell activating signals. Using a combination of in depth experimental analysis and mathematical modeling, at both the cellular and molecular levels, I hope to elucidate the homeostatic and dynamic regulation of the NF-kB signaling system in B-cell physiology.
Sun, S., J. Almaden, et al. (2005). “Assay development and data analysis of receptor-ligand binding based on scintillation proximity assay.” Metab Eng 7(1): 38-44.
Humphries, P. S., J. V. Almaden, et al. (2006). “Pyridine-2-propanoic acids: Discovery of dual PPARalpha/gamma agonists as antidiabetic agents.” Bioorg Med Chem Lett 16(23): 6116-9.
Humphries, P. S., S. Bailey, et al. (2006). “Pyridine-3-propanoic acids: Discovery of dual PPARalpha/gamma agonists as antidiabetic agents.” Bioorg Med Chem Lett 16(23): 6120-3.
Ralph, E. C., J. Almaden, et al. (2008). “Glucose modulation of glucokinase activation by small molecules.” Biochemistry 47(17): 5028-36.